We are interested in understanding how sex chromosomes have repeatedly evolved across the tree of life. We use stickleback fish as a model system to explore the evolutionary forces that shape sex chromosomes at the earliest stages of evolution. We combine molecular genetics, developmental genetics, and bioinformatic approaches to understand key processes in the formation of sex chromosomes, including the genetics of sex determination and the regulation of meiotic recombination.
1. Genetic mechanisms of sex determination
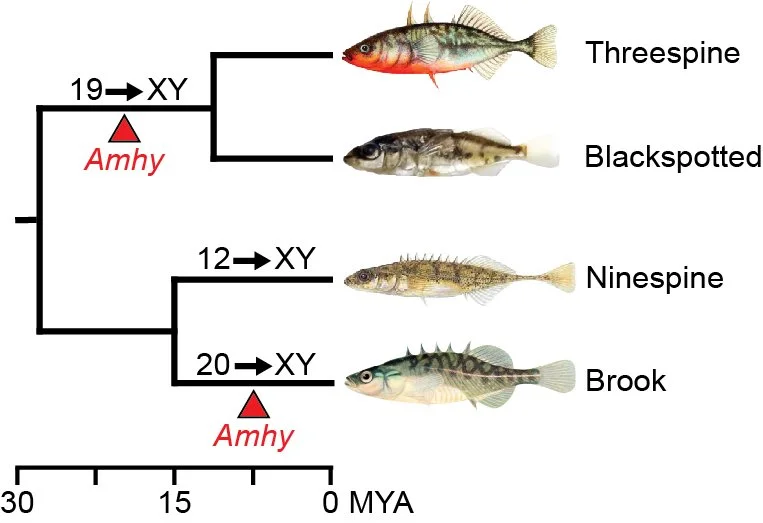
Threespine stickleback fish have a recently derived XY sex determination system, where sex is genetically determined. The key Y chromosome-linked sex determination gene remains to be identified. We are interested in understanding what genes on the Y chromosome are involved in sex determination as well as the underlying genes across the genome involved in the differentiation of sex. There is a tremendous diversity of sex determination mechanisms among animals and the closely related species of stickleback fish are not an exception. Multiple species of stickleback fish have independently derived sex chromosomes. We are interested in how sex determination occurs in different stickleback species to more broadly understand the plasticity of sex determination observed across animals.
2. Sex-specific differences in meiotic recombination
We are interested in understanding the mechanisms leading to differences in meiotic recombination between sexes. In species with heteromorphic sex chromosomes, recombination rates can vary widely across the X/Y or Z/W. In some regions of the chromosomes, recombination rates are orders of magnitude above genome-wide averages, whereas other regions experience complete suppression of crossing over between the chromosomes. We are exploring multiple stages in this process, including how double strand breaks are initiated across sex chromosomes and how double strand breaks are repaired. We are also interested in understanding why recombination rates can vary between sexes even across autosomes. To understand this, we combine molecular cytogenetics, genomics, and functional genetics experiments.